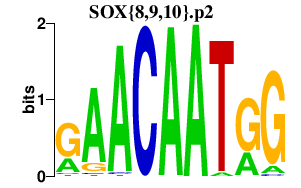
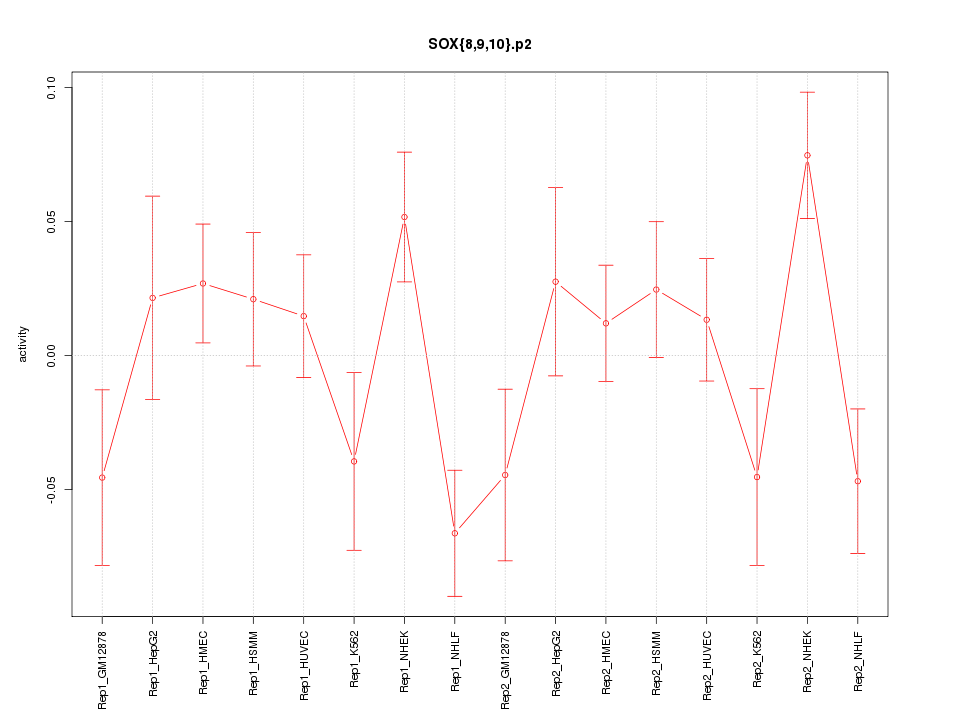
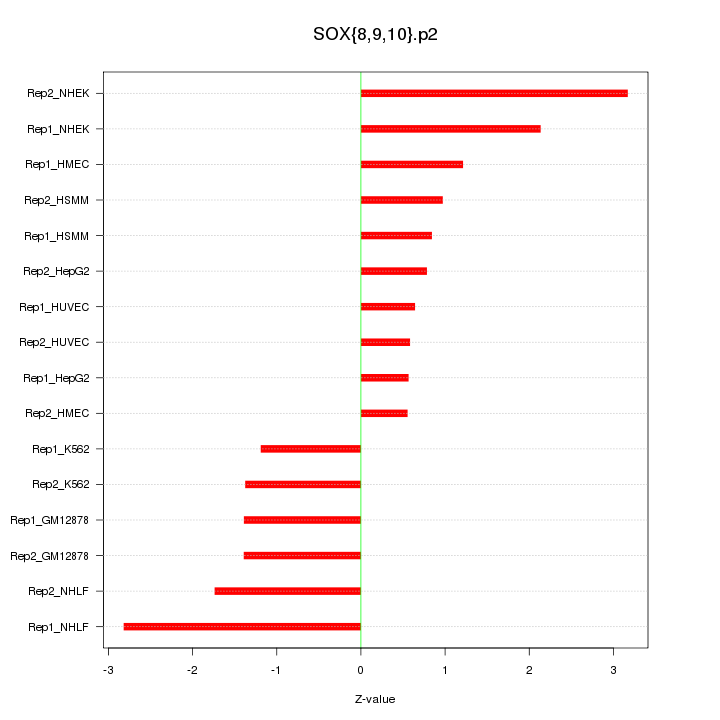
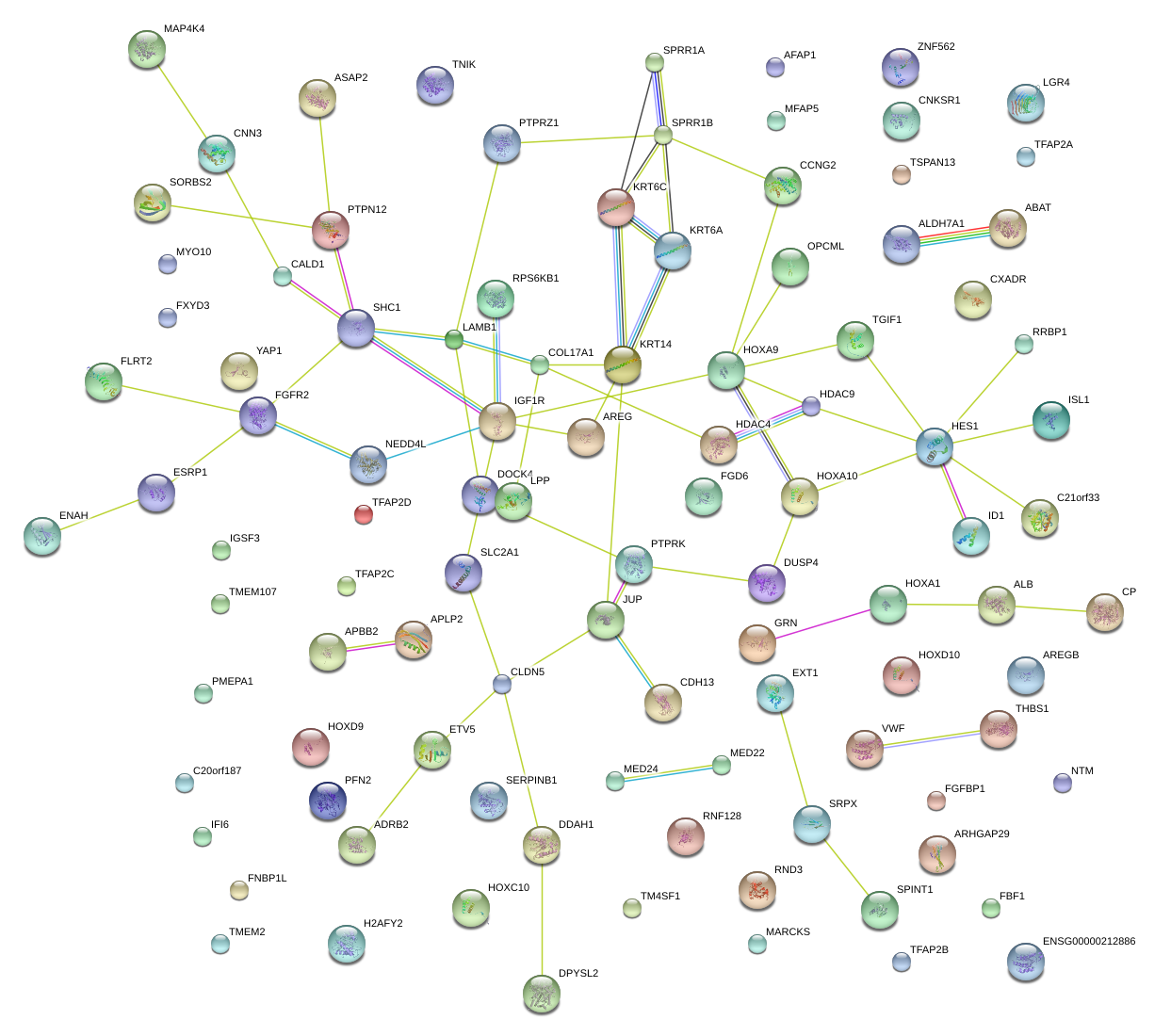
|
chr6_-_128883125
|
4.429
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr7_-_27171673
|
3.545
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr12_-_8706533
|
3.494
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr7_-_27171648
|
3.252
|
|
HOXA9
|
homeobox A9
|
|
chr17_-_36996611
|
3.038
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr12_-_51173238
|
2.619
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr12_-_8706637
|
2.529
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr2_+_101680594
|
2.489
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr21_+_17807167
|
2.160
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr11_-_27450808
|
1.964
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr1_-_43196911
|
1.923
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr5_-_16989371
|
1.911
|
NM_012334
|
MYO10
|
myosin X
|
|
chr1_-_94475504
|
1.878
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr12_+_52665196
|
1.727
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr7_+_134114957
|
1.711
|
|
CALD1
|
caldesmon 1
|
|
chr1_-_95165037
|
1.704
|
|
CNN3
|
calponin 3, acidic
|
|
chr7_+_121300394
|
1.633
|
NM_002851
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr10_+_71482362
|
1.594
|
NM_018649
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr10_+_71482542
|
1.571
|
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr4_-_186969041
|
1.570
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_-_105835609
|
1.556
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr1_+_151270301
|
1.509
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr1_-_117011824
|
1.508
|
NM_001007237
NM_001542
|
IGSF3
|
immunoglobulin superfamily, member 3
|
|
chr8_-_29262240
|
1.454
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr16_+_81218074
|
1.426
|
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr12_-_8706746
|
1.415
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr3_-_187309442
|
1.388
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chrX_-_37964924
|
1.384
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr3_-_151171576
|
1.371
|
|
PFN2
|
profilin 2
|
|
chr12_-_94135154
|
1.368
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr6_-_10523424
|
1.270
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr1_-_43197123
|
1.269
|
|
SLC2A1
|
solute carrier family 2 (facilitated glucose transporter), member 1
|
|
chr10_-_105835525
|
1.266
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr9_-_73573121
|
1.236
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr4_+_74491272
|
1.219
|
|
ALB
|
albumin
|
|
chr1_+_93686154
|
1.202
|
NM_001024948
NM_001164473
NM_017737
|
FNBP1L
|
formin binding protein 1-like
|
|
chr4_-_186969202
|
1.178
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_+_129496792
|
1.170
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr5_+_50714996
|
1.145
|
|
ISL1
|
ISL LIM homeobox 1
|
|
chr20_-_55718982
|
1.141
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr5_+_148186348
|
1.141
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr17_+_39778191
|
1.116
|
|
GRN
|
granulin
|
|
chr17_-_37196463
|
1.105
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr2_-_151052391
|
1.099
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr3_-_150578023
|
1.086
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr8_-_119192985
|
1.084
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr7_+_16759845
|
1.078
|
NM_014399
|
TSPAN13
|
tetraspanin 13
|
|
chr20_-_17610828
|
1.076
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr14_+_85066207
|
1.066
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr4_-_7991992
|
1.051
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr7_-_107430566
|
1.045
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr11_+_101488380
|
1.032
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr18_+_3439972
|
1.015
|
NM_173209
NM_173208
NM_003244
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr17_+_39778180
|
1.015
|
|
GRN
|
granulin
|
|
chr1_-_223907283
|
1.013
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr4_-_40631397
|
1.012
|
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr14_+_85066265
|
0.993
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr4_+_78297356
|
0.986
|
NM_004354
|
CCNG2
|
cyclin G2
|
|
chr1_-_85703198
|
0.969
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr18_+_3437607
|
0.961
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr1_-_94475840
|
0.958
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr1_-_153213454
|
0.955
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr4_+_75529716
|
0.951
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr4_-_15549047
|
0.947
|
NM_005130
|
FGFBP1
|
fibroblast growth factor binding protein 1
|
|
chr15_+_37660568
|
0.946
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr1_-_95165089
|
0.943
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr18_+_3441589
|
0.941
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr16_-_30841938
|
0.940
|
|
NCRNA00095
|
non-protein coding RNA 95
|
|
chrX_+_105823723
|
0.938
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr17_+_39778153
|
0.938
|
|
GRN
|
granulin
|
|
chr6_+_114285209
|
0.925
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr16_+_8714326
|
0.914
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr8_+_26491317
|
0.910
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr3_+_195336625
|
0.910
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr18_+_53862386
|
0.894
|
NM_001144967
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr22_-_17892859
|
0.891
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr15_+_38923914
|
0.887
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr17_-_35464137
|
0.886
|
|
MED24
|
mediator complex subunit 24
|
|
chr7_+_77005287
|
0.885
|
NM_001131008
NM_001131009
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr1_-_153213483
|
0.877
|
NM_001130041
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr18_+_54039779
|
0.871
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr8_+_95722516
|
0.868
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr2_+_9264360
|
0.857
|
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr8_+_95722575
|
0.849
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr1_-_153213425
|
0.847
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr2_-_239987557
|
0.847
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr4_+_78297418
|
0.839
|
|
CCNG2
|
cyclin G2
|
|
chr7_-_27102049
|
0.822
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr7_-_27186400
|
0.819
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr1_-_27871306
|
0.817
|
NM_002038
NM_022872
NM_022873
|
IFI6
|
interferon, alpha-inducible protein 6
|
|
chr7_+_18501876
|
0.805
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr3_-_150422474
|
0.804
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr5_-_125958503
|
0.800
|
NM_001182
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr12_-_6103945
|
0.798
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr18_+_54039818
|
0.795
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr20_+_29656744
|
0.795
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr10_-_123346148
|
0.792
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr17_-_1566223
|
0.778
|
|
C17orf91
|
chromosome 17 open reading frame 91
|
|
chr15_+_97009194
|
0.774
|
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr11_+_131285747
|
0.772
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr5_+_50714714
|
0.769
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr7_-_111633620
|
0.766
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr6_-_2785682
|
0.758
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr4_+_78298326
|
0.758
|
|
CCNG2
|
cyclin G2
|
|
chr1_+_26376560
|
0.754
|
NM_006314
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr3_+_189354167
|
0.753
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr19_+_40298571
|
0.734
|
NM_001136007
NM_001136008
NM_001136009
NM_001136010
NM_001136011
NM_001136012
NM_005971
NM_021910
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr3_+_51397731
|
0.729
|
NM_006010
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr6_-_11340886
|
0.728
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr1_-_68470585
|
0.719
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr14_+_58725104
|
0.717
|
NM_014992
|
DAAM1
|
dishevelled associated activator of morphogenesis 1
|
|
chr3_+_51397756
|
0.715
|
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr18_+_11741470
|
0.714
|
NM_001142339
NM_002071
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr8_+_102573837
|
0.714
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr20_+_60918836
|
0.713
|
NM_001853
|
COL9A3
|
collagen, type IX, alpha 3
|
|
chr8_-_81155564
|
0.700
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr5_+_40715778
|
0.699
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr1_+_167342149
|
0.693
|
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr17_+_43480674
|
0.685
|
NM_003204
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr3_+_195336631
|
0.685
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr4_+_146623405
|
0.674
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr20_-_17610704
|
0.671
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr3_+_189425886
|
0.664
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr16_-_88084387
|
0.662
|
NM_013275
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chr6_+_121798443
|
0.660
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr10_-_97311084
|
0.644
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr18_+_54039730
|
0.642
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr17_+_43480723
|
0.642
|
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr18_-_72857665
|
0.633
|
|
MBP
|
myelin basic protein
|
|
chr19_+_38556414
|
0.630
|
NM_001806
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma
|
|
chr11_+_60448487
|
0.625
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chr10_+_73393998
|
0.623
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr9_-_74757735
|
0.618
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr8_+_26491383
|
0.608
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr10_+_73703682
|
0.606
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr10_+_5126589
|
0.606
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr15_+_38923470
|
0.602
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr12_-_31370387
|
0.597
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr15_-_80428760
|
0.595
|
NM_001164465
|
GOLGA6L10
|
golgin A6 family-like 10
|
|
chr6_-_134680888
|
0.582
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr10_+_5126556
|
0.581
|
NM_003739
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr1_+_85819004
|
0.578
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr2_+_9264327
|
0.576
|
NM_001135191
NM_003887
|
ASAP2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr12_-_31370351
|
0.575
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr16_-_21344158
|
0.572
|
NM_130464
|
NPIPL3
|
nuclear pore complex interacting protein-like 3
|
|
chr9_-_14388981
|
0.571
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr5_+_74668748
|
0.571
|
NM_000859
NM_001130996
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase
|
|
chr3_-_140741221
|
0.570
|
NM_001130992
NM_001130993
NM_002899
|
RBP1
|
retinol binding protein 1, cellular
|
|
chr9_-_35101534
|
0.569
|
|
KIAA1539
|
KIAA1539
|
|
chr10_+_5126613
|
0.569
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr11_-_116213511
|
0.562
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr16_+_22432344
|
0.560
|
NM_001135865
|
LOC100132247
NPIPL3
|
nuclear pore complex interacting protein related gene
nuclear pore complex interacting protein-like 3
|
|
chr10_-_33287184
|
0.555
|
NM_002211
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr8_+_94998258
|
0.555
|
NM_018444
NM_001161780
NM_001161779
NM_001161781
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr12_-_31370305
|
0.553
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr17_-_35464414
|
0.551
|
NM_001079518
NM_014815
|
MED24
|
mediator complex subunit 24
|
|
chr11_-_74914514
|
0.547
|
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5
|
|
chr14_+_23008742
|
0.547
|
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr10_+_102096761
|
0.545
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr16_-_4927057
|
0.542
|
NM_002705
|
PPL
|
periplakin
|
|
chr3_-_101077605
|
0.539
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr1_-_203557379
|
0.538
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr1_+_109221124
|
0.535
|
NM_013296
|
GPSM2
|
G-protein signaling modulator 2
|
|
chrX_+_102498035
|
0.534
|
NM_001006612
NM_001006613
NM_001006614
NM_016303
|
WBP5
|
WW domain binding protein 5
|
|
chr8_+_95722448
|
0.534
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr1_+_109221271
|
0.530
|
|
GPSM2
|
G-protein signaling modulator 2
|
|
chr7_+_134114912
|
0.529
|
|
CALD1
|
caldesmon 1
|
|
chr11_+_13255836
|
0.529
|
NM_001030272
NM_001030273
NM_001178
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr2_-_39517633
|
0.528
|
NM_003618
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr3_+_190831909
|
0.522
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chr1_+_198975227
|
0.520
|
NM_203459
|
CAMSAP1L1
|
calmodulin regulated spectrin-associated protein 1-like 1
|
|
chr2_+_64534774
|
0.515
|
NM_014181
|
HSPC159
|
galectin-related protein
|
|
chr11_-_66482366
|
0.512
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr3_-_57558120
|
0.510
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr11_+_13255913
|
0.510
|
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr19_-_60352369
|
0.507
|
NM_001126132
NM_001126133
NM_003283
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr14_+_23008723
|
0.506
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr3_-_57558130
|
0.505
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr16_+_28869628
|
0.504
|
NM_032815
|
NFATC2IP
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein
|
|
chr4_-_187114407
|
0.504
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr7_-_41709191
|
0.503
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr17_+_43480724
|
0.499
|
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr10_+_124211353
|
0.497
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr3_-_11598829
|
0.494
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr12_-_10142779
|
0.492
|
NM_016511
|
CLEC1A
|
C-type lectin domain family 1, member A
|
|
chr2_-_160765008
|
0.492
|
|
ITGB6
|
integrin, beta 6
|
|
chr2_-_131838200
|
0.492
|
NM_001077637
|
LOC150786
|
RAB6C-like
|
|
chr1_+_67923660
|
0.491
|
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr6_+_158653679
|
0.487
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr16_+_64958025
|
0.484
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr20_-_45418818
|
0.484
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chrX_+_149902415
|
0.484
|
NM_005342
|
HMGB3
|
high-mobility group box 3
|
|
chr3_-_51976428
|
0.477
|
NM_020418
NM_001174100
NM_033008
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr1_-_150564302
|
0.474
|
NM_002016
|
FLG
|
filaggrin
|
|
chr22_+_31527678
|
0.473
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr16_-_83095720
|
0.473
|
NM_020947
|
KIAA1609
|
KIAA1609
|
|
chr11_-_128567302
|
0.469
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr22_+_36472262
|
0.466
|
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr13_-_40138607
|
0.466
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr3_-_57557906
|
0.466
|
|
ARF4
|
ADP-ribosylation factor 4
|